Viral interactions with the cytoskeleton in the manipulation of host cell biology
-
Project Leader

Dr Greg Moseleygregory.moseley@unimelb.edu.au
+61 3 8344 2288
Project Details
Using live-cell imaging and super resolution microscopy (Figure 1), immune signaling assays and viral reverse genetics/in vivo disease models, we have identified novel interactions of viral proteins with the microtubule (MT) cytoskeleton, whereby viruses can modulate the structure/dynamics of MTs to exploit their functions in regulating diverse processes including immunity, cell cycle progression, and apoptosis.
By generating mutant viruses unable to modulate MT-dependent functions we identified significant roles in virulence in vivo. Using these techniques, combined with functional genomics/proteomics, EM, and imaging techniques such as RICS and bimolecular complementation, current projects aim to understand the mechanisms underlying these effects, and to determine their potential as new targets to develop antiviral drugs/vaccines.
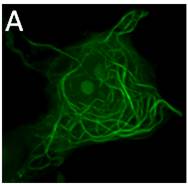
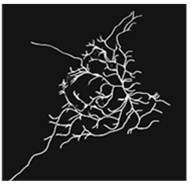
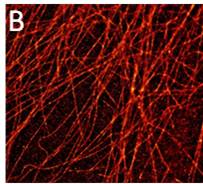
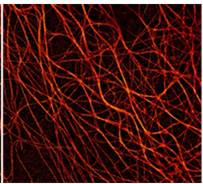
Figure 1: (A) 3D-MT association assay showing CLSM image (left) and automated tracing (right) of viral protein-MT interactions in living cells. (B) Super-resolution (dSTORM) microscopy reveals significant changes in MT structure between control (left) and viral protein-expressing cells (right).
Research Group
Faculty Research Themes
School Research Themes
Key Contact
For further information about this research, please contact the research group leader.
Department / Centre
Biochemistry and Molecular Biology
MDHS Research library
Explore by researcher, school, project or topic.